Reinforcement Learning
I’m interested in developing reinforcement learning (RL) methods that can adapt to challenging real-data settings such as healthcare. In particular I’m interested in offline RL for learning optimal policies in environments where direct exploration is expensive or unfeasible, settings when the Markovian assumption is violated such as in the dynamical treament regimen setting, where all medical history is relevant, and when not all rewards are observed which is often the case in electronic health records.
Expert-Supervised Reinforcement Learning for Offline Policy Learning and Evaluation, Aaron Sonabend W., Junwei Lu, Leo A. Celi, Tianxi Cai, Peter Szolovits. NeurIPS 2020. (pdf, code, video).
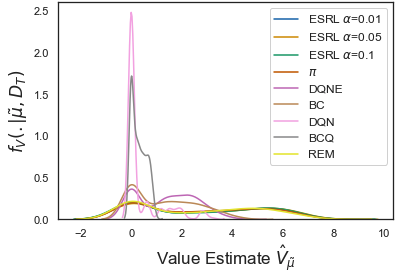
We develop an RL method which uses uncertainty quantification for offline policy learning. 1) the method can learn safe and optimal policies through hypothesis testing, 2) allows for different levels of risk aversion within the application context, and 3) yields interpretable acxtions at every state through posterior distributions. We also propose using this framework to compute off-policy value function posteriors. We provide theoretical guarantees for our estimators and regret bounds.
Semi-Supervised Off Policy Reinforcement Learning, Aaron Sonabend W., Nilanjana Laha, Rajarshi Mukherjee, Tianxi Cai. (pdf, 2020).
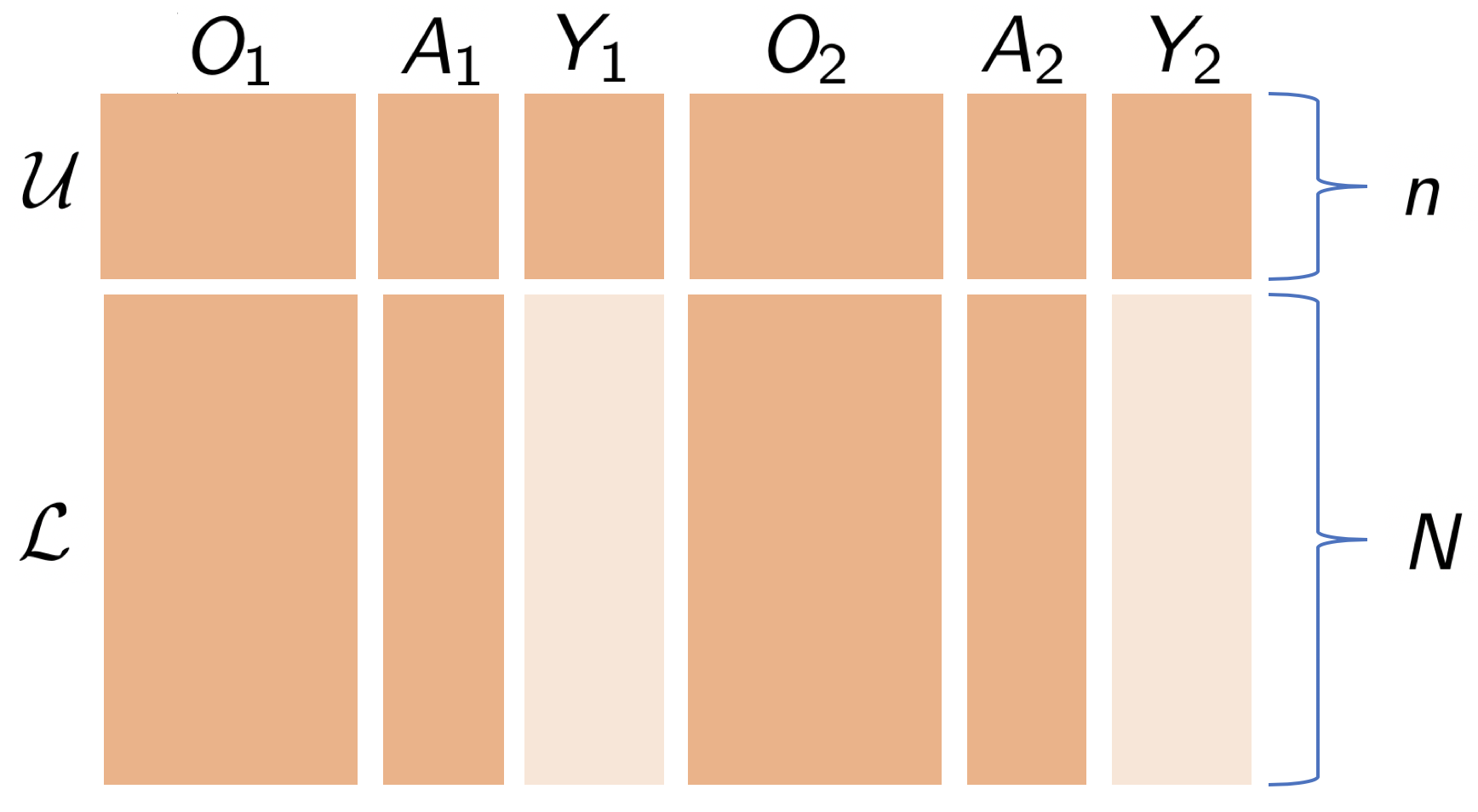
A common challenge with EHR data is that health-outcome information is often not well coded but rather embedded in clinical notes. Extracting precise outcome information often requires manual chart review, which is a resource intensive task. This translates into only having available small well-annotated cohorts. We develop a semi-supervised learning (SSL) approach that can efficiently leverage a small-sized labeled data with true outcome observed, and a large-sized unlabeled data with outcome surrogates observed. These surrogates are terms correlated to the outcome which can be extracted quickly from the clinical notes using NLP tools. In order to do this, we propose a theoretically justified SSL approach to Q-learning, and develop a robust and efficient SSL approach to estimating the value function, defined as the expected counterfactual outcome under the optimal sequential treatment regime (STR). We provide theoretical results to understand when and to what degree efficiency can be gained from our procedure, which in the worst case is as efficient as using only labeled data. Our approach is imputation based, and it is robust to miss-specification of our imputation models. Further, we provide a doubly robust value function estimator for the derived STR which is asymptotically normal. If either the Q-functions or the propensity score models are correctly specified, our estimator is root n-consistent for the true counterfactual value. We use our method to find the optimal STR for inflammatory bowel disease.
Surrogate loss function for learning STRs, Nilanjana Laha, Aaron Sonabend W., Rajarshi Mukherjee, Tianxi Cai. (Work in progress).
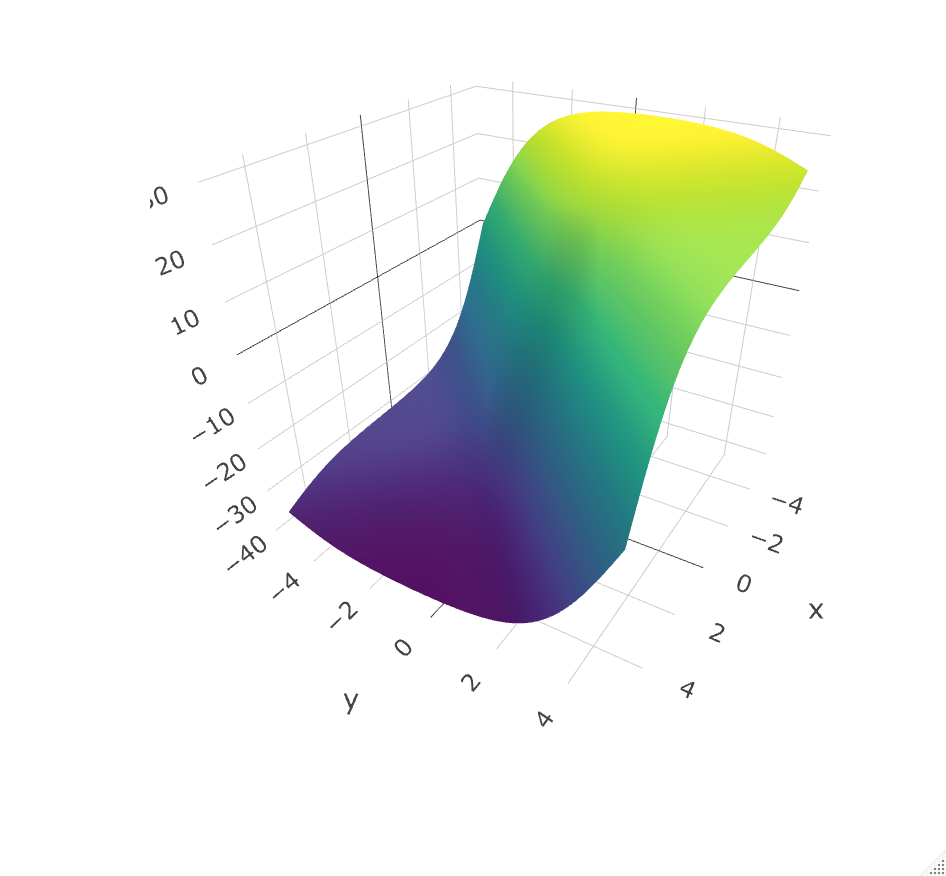
Offline RL methods seek to find the policy which maximizes the counterfactual value function. Direct optimization of such value is challenging however, as this function is non-smooth and discontinuous. This problem is often addressed through indirect methods such as Q-learning, which rely on correct specification of the conditional mean model. An alternative in the single time-point case is to recast the value optimization into a classification problem; propose a decision function; then use a smooth, concave surrogate value function to approximate the value of interest. This surrogate allows us to directly maximize a nonparametric estimator of the objective over a large class of functions. Surrogates that satisfy simple conditions have been shown to be Fisher consistent. Interestingly, when generalizing to multiple time-points, concave surrogate functions are not Fisher consistent. We explore this problem, illustrate why a simple concave generalization fails, and propose a class of smooth surrogate functions which achieve Fisher consistency. We derive regret bounds for the optimal STR under this class of surrogates, when using neural networks or wavelets to learn $f$. With the proposed smooth surrogates, our approach scales with sample size as opposed to alternative approaches that, similar to support vector machines, optimize in the dual space.
Phenotyping with Natural Language Processing
I’m interested in using NLP for automatic diagnosing, this is a hard problem where usually there is a lack of labels, therefore unsupervisde phenotyping methods such as UNITE are highly attractive and generalize well across ddatasets. Additionally, I’m interested in using natural language to discover patters behind psychiatric behavior.
Automated ICD coding via unsupervised knowledge integration (UNITE), Aaron Sonabend W., Winston Cai, Yuri Ahuja, Ashwin Ananthakrishnan, Zongqi Xia, Sheng Yu, Chuan Hong. International Journal of Medical Informatics, 2020.
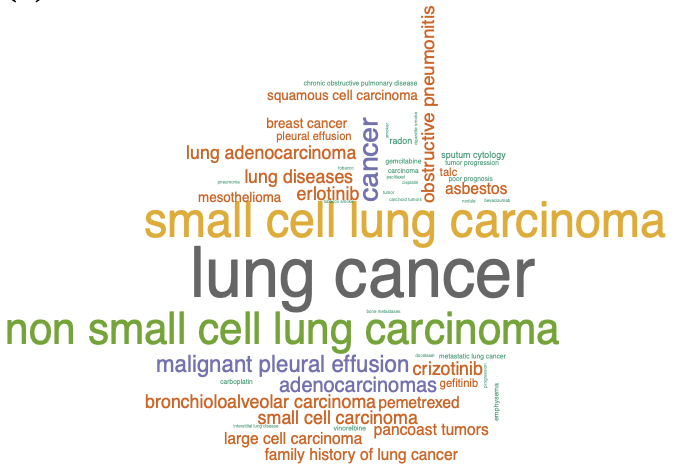
We developed an unsupervised knowledge integration (UNITE) algorithm to automatically assign phenotype codes for a specific disease by analyzing clinical narrative notes via semantic relevance assessment. UNITE has substantially better performance than unsupervised methods like topic models, and the performance is consistent across different electronic medical records (EMR) systems, which is often an issue with deep learning models. It also requires no human annotation of labels which makes it robust to bias often found within EMR systems.
Integrating questionnaire measures for transdiagnostic psychiatric phenotyping using word2vec, Aaron Sonabend W., Amelia M. Pellegrini, Stephanie Chan, Hannah E. Brown, James N. Rosenquist, Pieter J. Vuijk, Alysa E. Doyle, Roy H. Perlis , Tianxi Cai. PLOS ONE, 2020.
We propose a method for applying natural language processing to derive dimensional measures of psychiatric symptoms from questionnaire data. We used 9 symptom measures from cellular biobanking study that enrolled individuals with mood and psychotic disorders, as well as healthy controls. A low-dimensional approximation to the embedding space was used to derive succinct summary of symptom profiles. To validate our embedding-based disease profiles, these were compared to presence or absence of axis I diagnoses derived from structured clinical interview, and to objective neurocognitive testing. We also found that unsupervised clustering of these low-dimensional profiles discriminate well between cases and controls. Our derived symptom measures and estimated Research Domain Criteria scores also associated significantly with performance on neurocognitive tests.
Scalable semiparametric Bayesian inference
Scalable Gaussian Process Regression Via Median Posterior Inference for Estimating Multi-Pollutant Mixture Health Effects, Aaron Sonabend W., Jiangshan Zhang, Joel Schwartz, Brent Coull, Junwei Lu. Machine Learning in Public Health Workshop, NeurIPS 2020.
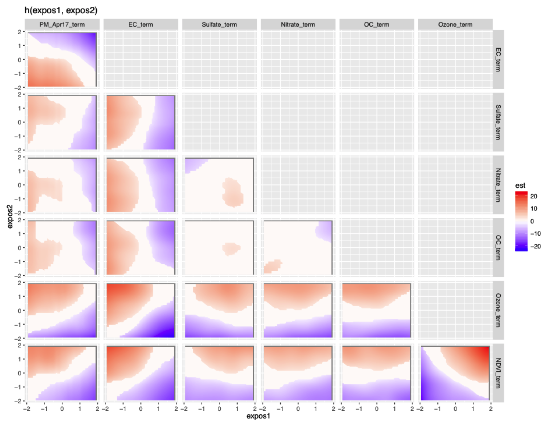
We come up with a divide-and-conquer semiparametric Bayesian inference algorithm to learn flexible models for the impact of pollution on public health. The relationship between health outcomes and multiple pollutants is highly complex and needs large data sets to be measured. On the other hand most Bayesian methodology used in this context is prohibitively slow with large samples. We extend a semi-parametric Gaussian process regression framework to scale to large data sets. We assume a Gaussian semi-paramteric relationship of the health ooutcomes, pollutants and confounders. We propose a sample-splitting approach to learn the Gaussian process for the health outcome-pollutant relationship for which we obtain a nonparametric convergence rate.